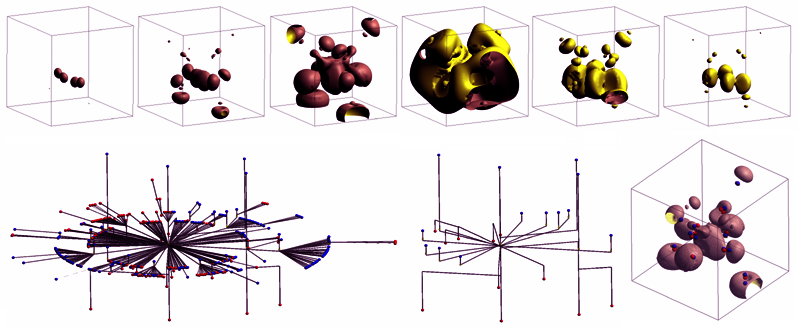
| left mouse button click & drag
| rotate the scene
|
| [SHIFT] + left mouse button click & drag (along the X axis)
| zoom in, zoom out the scene
|
| middle mouse button click & drag
| change the pan of the actual scene
|
| [left] [right] keyboard arrows
|
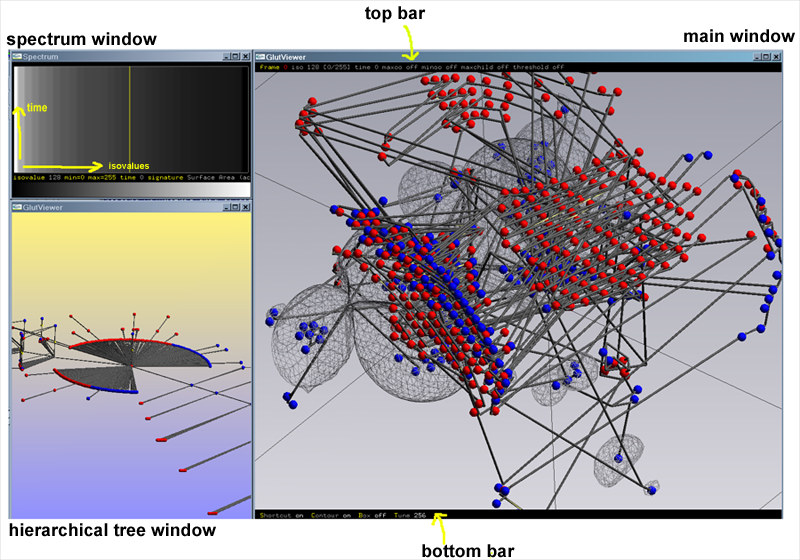
Switch for the change of a specific value:
(see highlighted item on the main window top bar):
frame, isovalue, time, max overall, min overall, max number of children, threshold
|
| [up] [down] keyboard arrows
|
change the current value, up increases it,
down decreases it
(to set the"sensibility" of the change use
't' and 'T')
|
| 'g'
|
change the view mode of the embedded contour three
|
| 'B'/'b'
| enable/disable or increase/decrease the size of the
bounding box which specifies the current region of
interest
|
| 'm'
| Only for triangular mesh. Change the view mode of the
current mesh
|
| 'c'
| enable/disable or change the view mode of the level sets
of the actual dataset
|
| 'R'/'r'
| increase/decrease the radius of the spheres of the contour
trees
|
| 'D'/'d'
| increase/decrease the radius of the cylinders of the
contour trees
|
| 's'
| save the current frame or enable/disable shortcut
|
| 'o'
| open the current frame if the highlighted item is "frame" in the top bar.
otherwise set to 'off' the filter for maximum overall, minimum overall,
threshold or max number of children
|
| left mouse button click & and drag
| rotate the scene
|
| [SHIFT] + left mouse button click & drag (along the X axis)
| zoom in, zoom out the scene
|
| middle mouse button click & drag
| change the pan of the actual scene
|
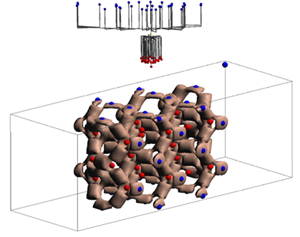
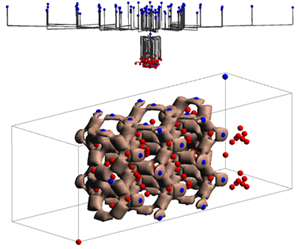
| 'g'
| change the view mode of the hierarchical contour three
|
| 'X'/'x'
| scale (increase or decrease) the space occupancy along the X axis
|
| 'Y'/'y'
| scale (increase or decrease) the space occupancy along the Y axis
|
| 'Z'/'z'
| scale (increase or decrease) the space occupancy along the Z axis
|